Full Plasma (Core + Edge profiles)¶
This notebook demonstrates how to load and visualize a plasma object produced by cherab’s Plasma class using the cherab.imas interface.
The example test data was calculated by JINTRAC for an ITER 15 MA H-mode scenario.
Prerequisites: Pooch must be installed to download the example data.
[1]:
import numpy as np
import ultraplot as uplt
from matplotlib.colors import SymLogNorm
from rich import print as rprint
from rich.table import Table
from cherab.core.math import sample3d, sample3d_grid
from cherab.imas.datasets import iter_jintrac
from cherab.imas.plasma import load_plasma
# Set dark background for plots
uplt.rc.style = "dark_background"
uplt.rc["title.borderwidth"] = 0
00:24:01 CRITICAL Could not import 'imas_core': No module named 'imas_core'. Some functionality is not available. @imas_interface.py:34
Define a function to plot plasma quantities¶
[2]:
def plot_quantity(
ax: uplt.axes.Axes,
quantity: np.ndarray,
extent: tuple[float, float, float, float],
logscale: bool = False,
symmetric: bool = False,
) -> uplt.axes.Axes:
"""Make a 2D plot of quantity, optionally on a log scale."""
if logscale:
# Plot lowest values (mainly 0's) on linear map, as log(0) = -inf.
linthresh = np.percentile(np.unique(quantity), 1)
norm = SymLogNorm(
linthresh=float(max(linthresh, 1.0e-10 * quantity.max())),
base=10,
)
else:
norm = None
# Sampled data is indexed as quantity(x, y),
# but matplotlib's imshow expects quantity(y, x).
if symmetric and not logscale:
vmax = np.abs(quantity).max()
image = ax.imshow(
quantity.T,
extent=extent,
discrete=False,
origin="lower",
vmin=-vmax,
vmax=vmax,
cmap="berlin",
)
else:
image = ax.imshow(
quantity.T,
extent=extent,
discrete=False,
origin="lower",
norm=norm,
cmap="gnuplot",
)
ax.colorbar(
image,
formatter="log",
tickminor=True,
)
ax.format(
aspect=1,
xlabel="$R$ [m]",
ylabel="$Z$ [m]",
xlocator=1,
ylocator=1,
)
return ax
Retrieve ITER JINTRAC sample data¶
[3]:
path = iter_jintrac()
Load the plasma object¶
The instance of the `Plasma <https://www.cherab.info/plasmas/core_plasma_classes.html#cherab.core.Plasma>`__ class is created by loading the core_profiles and edge_profiles IDSs from the IMAS database.
The equilibrium information is automatically loaded from the equilibrium IDS if not already provided.
[4]:
plasma = load_plasma(path, "r")
00:24:02 INFO Parsing data dictionary version 4.1.1 @dd_zip.py:89
00:24:02 INFO Parsing data dictionary version 4.0.0 @dd_zip.py:89
/home/runner/work/imas/imas/src/cherab/imas/plasma/blend.py:140: RuntimeWarning: The 'get_slice' method is not implemented for the URI '/home/runner/.cache/cherab/imas/iter_scenario_53298_seq1_DD4.nc'. Falling back to 'get' method to retrieve the entire IDS.
core_profiles_ids = get_ids_time_slice(
/home/runner/work/imas/imas/src/cherab/imas/plasma/blend.py:162: RuntimeWarning: The 'get_slice' method is not implemented for the URI '/home/runner/.cache/cherab/imas/iter_scenario_53298_seq1_DD4.nc'. Falling back to 'get' method to retrieve the entire IDS.
edge_profiles_ids = get_ids_time_slice(
/home/runner/work/imas/imas/src/cherab/imas/plasma/equilibrium.py:96: RuntimeWarning: The 'get_slice' method is not implemented for the URI '/home/runner/.cache/cherab/imas/iter_scenario_53298_seq1_DD4.nc'. Falling back to 'get' method to retrieve the entire IDS.
equilibrium_ids = get_ids_time_slice(
/home/runner/work/imas/imas/src/cherab/imas/plasma/equilibrium.py:178: RuntimeWarning: The 'get_slice' method is not implemented for the URI '/home/runner/.cache/cherab/imas/iter_scenario_53298_seq1_DD4.nc'. Falling back to 'get' method to retrieve the entire IDS.
equilibrium_ids = get_ids_time_slice(
Warning! Unable to verify that the cell nodes are in the winding order.
Warning! Using average ion temperature for the DT ion.
Warning! Using average ion temperature for the He+1 ion.
Warning! Using average ion temperature for the He+2 ion.
Warning! Using average ion temperature for the Ne(bundle 1) ion.
Warning! Using average ion temperature for the Ne(bundle 5) ion.
Warning! Using average ion temperature for the W(bundle 1) ion.
Warning! Using average ion temperature for the W(bundle 6) ion.
Warning! Using average ion temperature for the Ne(bundle 2) ion_bundle.
Warning! Using average ion temperature for the Ne(bundle 3) ion_bundle.
Warning! Using average ion temperature for the Ne(bundle 4) ion_bundle.
Warning! Using average ion temperature for the W(bundle 2) ion_bundle.
Warning! Using average ion temperature for the W(bundle 3) ion_bundle.
Warning! Using average ion temperature for the W(bundle 4) ion_bundle.
Warning! Using average ion temperature for the W(bundle 5) ion_bundle.
Warning! Species of type 'molecule' are currently not supported. The following species will be skipped:
DT2
Warning! Species of type 'ion_bundle' are currently not supported. The following species will be skipped:
; ; ; ; ; ; ; ; ;
Warning! Species of type 'ion_bundle' are currently not supported. The following species will be skipped:
Ne(bundle 2); Ne(bundle 3); Ne(bundle 4); W(bundle 2); W(bundle 3); W(bundle 4); W(bundle 5)
The loaded plasma object contains both core and edge profiles and all available species information shown below.
[5]:
# Organize plasma species by symbol and charge
species = {}
for s in plasma.composition:
symbol = s.element.symbol
if symbol not in species:
species[symbol] = []
species[symbol].append(s)
# Sort species of each element by charge
for symbol in species:
species[symbol].sort(key=lambda x: x.charge)
# Print table of plasma species
table = Table(title="Plasma Species")
table.add_column("Element", style="cyan", justify="right")
table.add_column("Charge")
for symbol in species:
for i, s in enumerate(species[symbol]):
if i == 0:
table.add_row(symbol, f"{s.charge:>2d}+")
else:
table.add_row("", f"{s.charge:>2d}+")
rprint(table)
Plasma Species ┏━━━━━━━━━┳━━━━━━━━┓ ┃ Element ┃ Charge ┃ ┡━━━━━━━━━╇━━━━━━━━┩ │ D │ 0+ │ │ │ 1+ │ │ T │ 0+ │ │ │ 1+ │ │ He │ 0+ │ │ │ 1+ │ │ │ 2+ │ │ W │ 0+ │ │ │ 1+ │ │ │ 74+ │ │ Ne │ 0+ │ │ │ 1+ │ │ │ 10+ │ └─────────┴────────┘
Plot several species’ profiles¶
Define some constants related to sampling and plotting.
[6]:
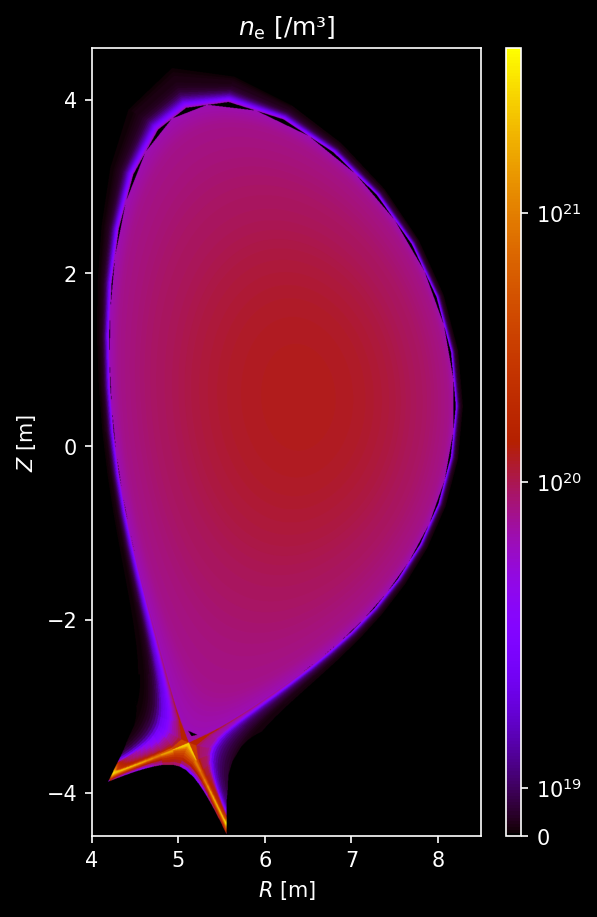
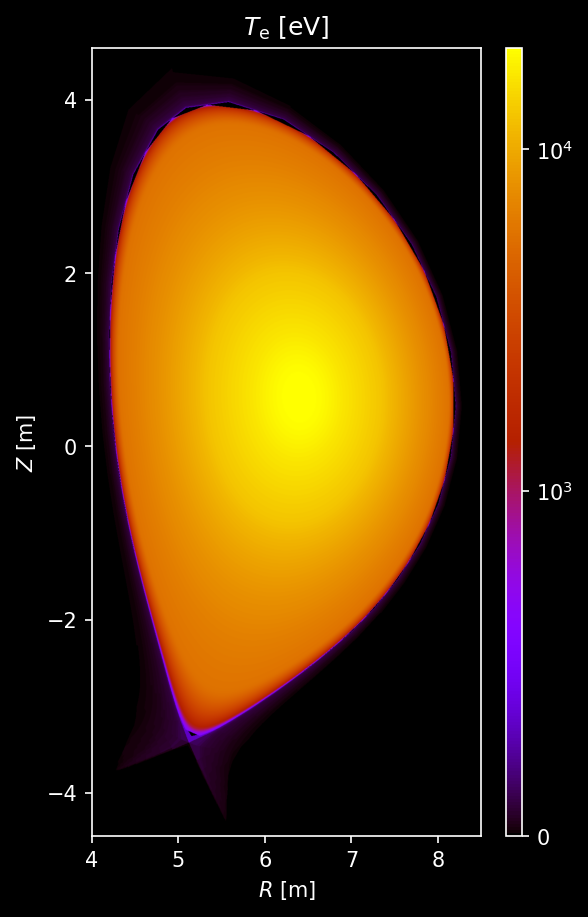
Electron density and temperature¶
[7]:
# Sample electron density
r_pts, _, z_pts, n_e = sample3d(
plasma.electron_distribution.density,
(R_MIN, R_MAX, n_r),
(0, 0, 1),
(Z_MIN, Z_MAX, n_z),
)
n_e = n_e.squeeze()
# Plot electron density
fig, ax = uplt.subplots()
ax = plot_quantity(
ax,
n_e,
extent,
logscale=True,
)
ax.format(
title="$n_\\mathrm{e}$ [/m³]",
)
# Sample electron temperature
te_plasma = sample3d_grid(
plasma.electron_distribution.effective_temperature,
r_pts,
[0],
z_pts,
).squeeze()
# Plot electron temperature
fig, ax = uplt.subplots()
ax = plot_quantity(
ax,
te_plasma,
extent,
logscale=True,
)
ax.format(
title="$T_\\mathrm{e}$ [eV]",
)
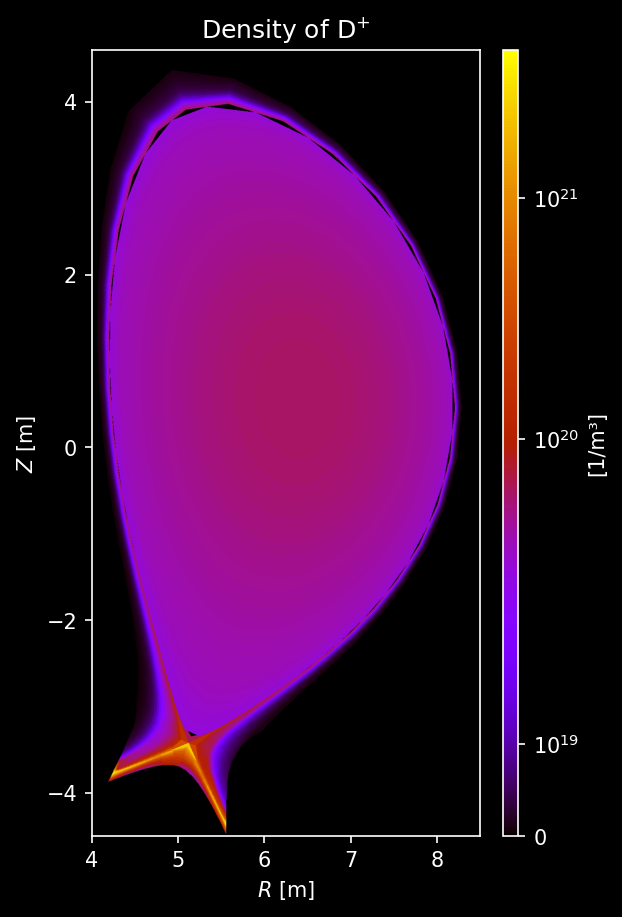
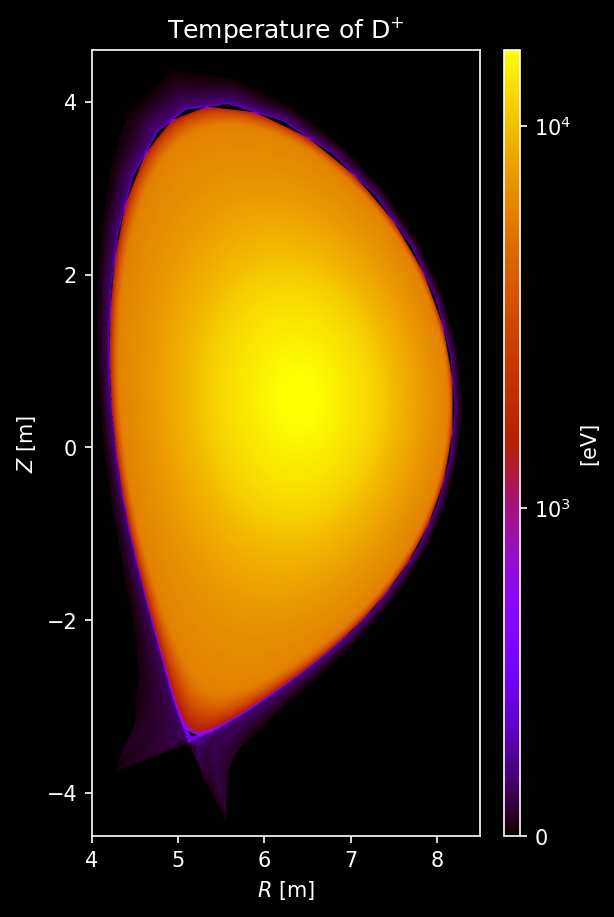
Hydrogenic species (D, T) density and temperature¶
[8]:
def species_label(species) -> str:
"""Construct element symbol with charge label"""
if species.charge == 0:
label = f"{species.element.symbol}$^{{0}}$"
elif species.charge == 1:
label = f"{species.element.symbol}$^{{+}}$"
else:
label = f"{species.element.symbol}$^{{{species.charge}+}}$"
return label
# === Density Plot ===
fig, axs = uplt.subplots(
nrows=2,
ncols=2,
)
axs.format(suptitle="Hydrogenic Species Density [/m³]")
for i_row, s_list in enumerate([species["D"], species["T"]]):
for i_col, s in enumerate(s_list):
# Sample density
density = sample3d_grid(
s.distribution.density,
r_pts,
[0],
z_pts,
).squeeze()
# Plot density
ax = plot_quantity(
axs[i_row, i_col],
density,
extent,
logscale=True,
)
ax.format(urtitle=species_label(s))
# === Temperature Plot ===
fig, axs = uplt.subplots(
nrows=2,
ncols=2,
)
axs.format(suptitle="Hydrogenic Species Temperature [eV]")
for i_row, s_list in enumerate([species["D"], species["T"]]):
for i_col, s in enumerate(s_list):
# Sample temperature
temperature = sample3d_grid(
s.distribution.effective_temperature,
r_pts,
[0],
z_pts,
).squeeze()
# Plot deutrium temperature
ax = plot_quantity(
axs[i_row, i_col],
temperature,
extent,
logscale=True,
)
ax.format(urtitle=species_label(s))
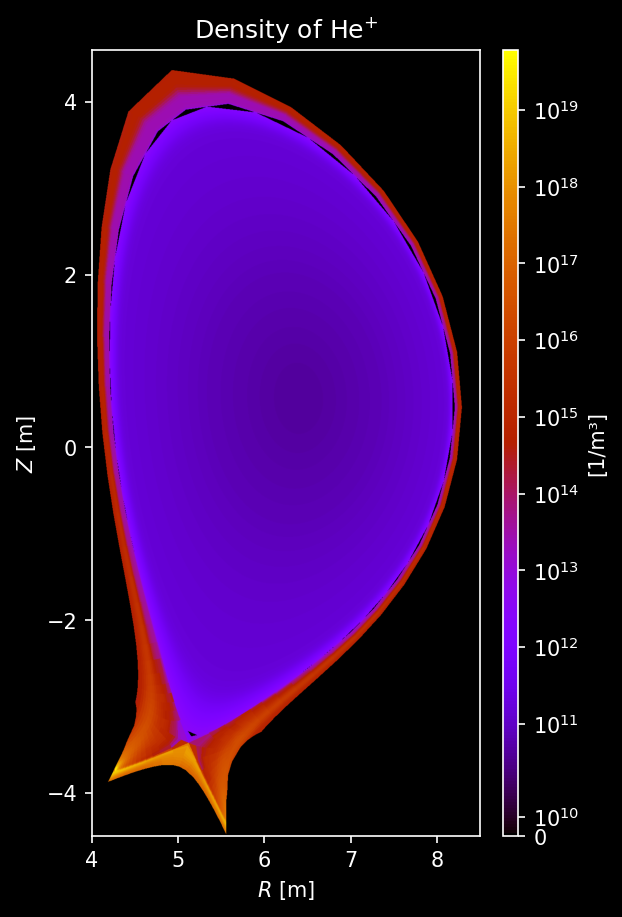
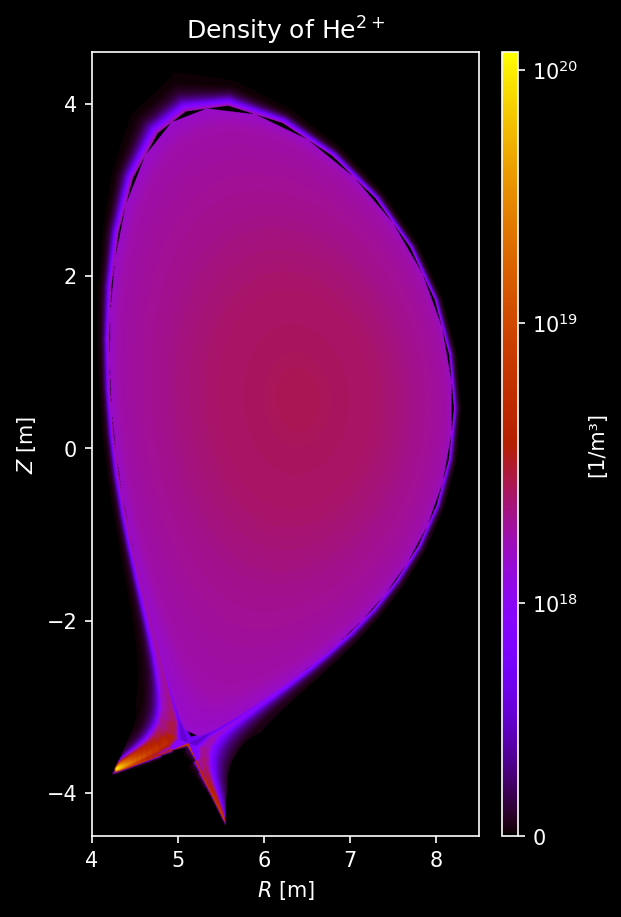
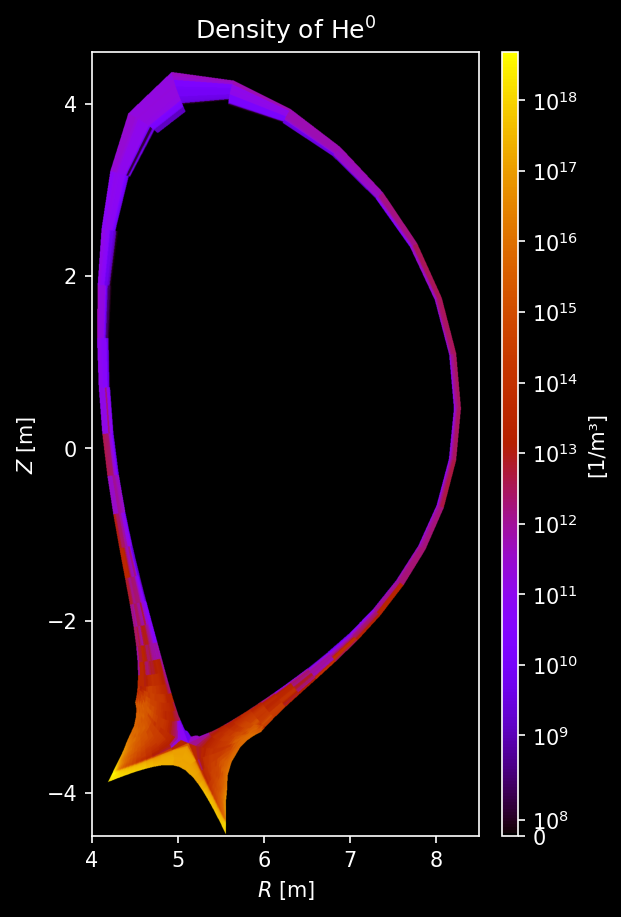
Other species’ density¶
[9]:
for s_list in [species["He"], species["Ne"], species["W"]]:
# Each element gets its own figure
fig, axs = uplt.subplots(
ncols=len(s_list),
)
axs.format(suptitle=f"{s_list[0].element.name.capitalize()} Density [/m³]")
for i_col, s in enumerate(s_list):
# Sample density
density = sample3d_grid(
s.distribution.density,
r_pts,
[0],
z_pts,
).squeeze()
# Make non-physical negative densities zero
density[density <= 0] = 0.0
# Plot density
ax = plot_quantity(
axs[i_col],
density,
extent,
logscale=True,
)
ax.format(urtitle=species_label(s))